Tasks#
|
|
Task configs are ‘high-level’ configs which configure the transforms outputs, losses and metrics of the model.
Specific training objects are achieved through the use of Transforms.
Note
To change the task, use a command with a format like:
workshop train task=<TASK_NAME> dataset=cath encoder=gvp trainer=cpu ...
# or
python proteinworkshop/train.py task=<TASK_NAME> dataset=cath encoder=gvp trainer=cpu ... # or trainer=gpu
Where <TASK_NAME> is one of the tasks listed below.
See also
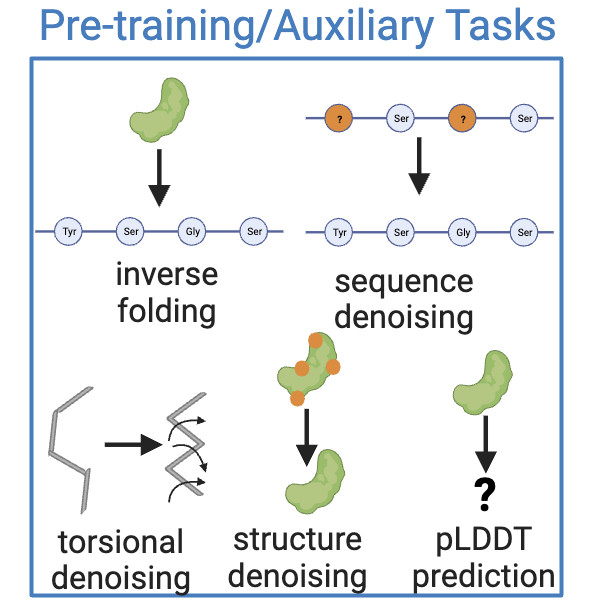
Denoising Tasks#
Sequence Denoising (sequence_denoising)#
This config trains a model to predict the original identies of the corrupted residues in a protein sequence.
The corruption is configured by the sequence noise transform <proteinworkshop.tasks.sequence_denoising.SequenceNoiseTransform>, which can be configured to apply masking or mutation
corruptions in various amounts to the input sequence.
config/task/sequence_denoising.yaml
# @package _global_
defaults:
- override /metrics:
- accuracy
- f1_score
- perplexity
- override /decoder:
- residue_type
- override /transforms:
- remove_missing_ca
- sequence_denoising
dataset:
num_classes: 23
callbacks:
early_stopping:
monitor: val/residue_type/accuracy
mode: "max"
model_checkpoint:
monitor: val/residue_type/accuracy
mode: "max"
task:
task: "sequence_denoising"
classification_type: "multiclass"
metric_average: "micro"
losses:
residue_type: cross_entropy
label_smoothing: 0.0
output:
- residue_type
supervise_on:
- residue_type
Structure Denoising (structure_denoising)#
This config trains a model to predict the original Cartesian coordinates of the corrupted residues in a protein structure.
Noise is applied to the Cartesian coordinates and the model is tasked with predicting either the per-residue Cartesian noise or the original Cartesian coordinates.
The corruption is configured by the structure noise transform <proteinworkshop.tasks.structure_denoising.StructureNoiseTransform>`, which can be configured to apply uniform or gaussian random noise in various amounts to the input structure.
See also
proteinworkshop.tasks.structure_denoising.StructureNoiseTransform
# @package _global_
defaults:
- override /metrics:
- rmse
- override /decoder:
- pos_equiv
- override /transforms:
- remove_missing_ca
- structure_denoising
dataset:
num_classes: null
callbacks:
early_stopping:
monitor: val/loss/pos
mode: "min"
model_checkpoint:
monitor: val/loss/pos
mode: "min"
task:
task: "structure_denoising"
losses:
pos: mse_loss
label_smoothing: 0.0
output:
- pos
supervise_on:
- pos
Sequence & Structure Denoising (sequence_structure_denoising)#
This config trains a model to predict the original identies of the corrupted residues in a protein sequence and the original Cartesian coordinates of the corrupted residues in a protein structure.
This config demonstrates how we can compose transforms in a modular fashion to create new training regimes.
See also
proteinworkshop.tasks.sequence_denoising.SequenceNoiseTransform
proteinworkshop.tasks.structure_denoising.StructureNoiseTransform
# @package _global_
defaults:
- override /metrics:
- rmse
- accuracy
- override /decoder:
- pos_equiv
- residue_type
- override /transforms:
- remove_missing_ca
- structure_denoising
- sequence_denoising
dataset:
num_classes: 23
callbacks:
early_stopping:
monitor: val/loss/total
mode: "min"
model_checkpoint:
monitor: val/loss/total
mode: "min"
task:
task: "sequence_structure_denoising"
classification_type: "multiclass"
metric_average: "micro"
losses:
pos: mse_loss
residue_type: cross_entropy
label_smoothing: 0.0
output:
- pos
- residue_type
supervise_on:
- pos
- residue_type
Torsional Denoising (torsional_denoising)#
This config trains a model to predict the original dihedral angles of the corrupted residues in a protein structure.
The torsional noise transform applies noise in dihedral angle space. The cartesian coordinates are the recomputed using pNeRF to enable structure-based featurisation.
# @package _global_
defaults:
- override /metrics:
- rmse
- override /decoder:
#- dihedrals
- torsional_noise
- override /transforms:
- remove_missing_ca
- torsional_denoising
dataset:
num_classes: null
callbacks:
early_stopping:
#monitor: val/loss/dihedrals
monitor: val/loss/torsional_noise
mode: "min"
model_checkpoint:
#monitor: val/loss/dihedrals
monitor: val/loss/torsional_noise
mode: "min"
task:
task: "torsional_denoising"
losses:
#dihedrals: mse_loss
torsional_noise: mse_loss
label_smoothing: 0.0
output:
#- dihedrals # Or torsional_noise
- torsional_noise # or dihedrals
supervise_on:
# - dihedrals
- torsional_noise
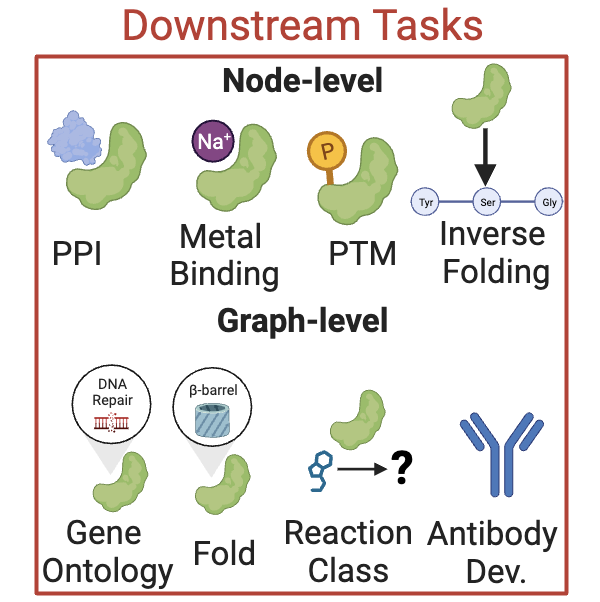
Node-level Tasks#
Protein-Protein Interaction Site Prediction (ppi_site_prediction)#
# @package _global_
defaults:
- _self_
- override /metrics:
- accuracy
- f1_score
- f1_max
- auprc
- rocauc
- override /decoder:
- node_label
- override /transforms:
- remove_missing_ca
- ppi_site_prediction
dataset:
num_classes: 2
callbacks:
early_stopping:
monitor: val/node_label/accuracy
mode: "max"
model_checkpoint:
monitor: val/node_label/accuracy
mode: "max"
task:
task: "classification"
classification_type: "binary"
metric_average: "micro"
losses:
node_label: bce
label_smoothing: 0.0
output:
- "node_label"
supervise_on:
- "node_label"
Ligand Binding Site Prediction (binding_site_identification)#
See also
proteinworkshop.tasks.binding_site_identification.BindingSiteIdentificationTransform
# @package _global_
defaults:
- override /metrics:
- accuracy
- f1_score
- auprc
- rocauc
- override /decoder:
- node_label
- override /transforms:
- remove_missing_ca
- binding_site_prediction
transforms:
binding_site_prediction:
hetatms: [HOH, SO4, PEG] # Types of hetatms to be considered as binding sites
threshold: 3.5 # Threshold for binding site prediction
ca_only: False # Whether to use only CA atoms for assigning labels
multilabel: True # Whether to use multilabel or binary labels
dataset:
num_classes: 3 # This needs to match the number of hetatms above
callbacks:
early_stopping:
monitor: val/node_label/accuracy
mode: "max"
model_checkpoint:
monitor: val/node_label/accuracy
mode: "max"
task:
task: "binding_site_identification"
classification_type: "multilabel" # Check this aligns with binding site config above
metric_average: "macro"
losses:
node_label: cross_entropy # Check this aligns with binding site config above
label_smoothing: 0.0
output:
- "node_label"
supervise_on:
- "node_label"
Multiclass Node Classification (multiclass_node_classification)#
# @package _global_
defaults:
- override /metrics:
- accuracy
- f1_score
- f1_max
- override /decoder:
- node_label
callbacks:
early_stopping:
monitor: val/node_label/accuracy
mode: "max"
model_checkpoint:
monitor: val/node_label/accuracy
mode: "max"
task:
task: "classification"
classification_type: "multiclass"
metric_average: "micro"
losses:
node_label: cross_entropy
label_smoothing: 0.0
output:
- "node_label"
supervise_on:
- "node_label"
pLDDT Prediction (plddt_prediction)#
This config specifies a self-supervision task to predict the per-residue pLDDT score of each node.
Warning
This task requires the input data to have a b_factor attribute.
If the input structure are not predicted structures, this task will be a B factor prediction task.
# @package _global_
defaults:
- override /metrics:
- rmse
- override /decoder:
- b_factor
dataset:
num_classes: 1
callbacks:
early_stopping:
monitor: val/loss/b_factor
mode: "min"
model_checkpoint:
monitor: val/loss/b_factor
mode: "min"
task:
task: "plddt_prediction"
losses:
b_factor: mse_loss
label_smoothing: 0.0
output:
- b_factor
supervise_on:
- b_factor
Edge-level Tasks#
Edge Distance Prediction (edge_distance_prediction)#
This config specifies a self-supervision task to predict the pairwise distance between two nodes.
We first sample num_samples edges randomly from the input batch. We then
construct a mask to remove the sampled edges from the batch. We store the
masked node indices and their pairwise distance as batch.node_mask and
batch.edge_distance_labels, respectively. Finally, it masks the edges
(and their attributes) using the constructed mask and returns the modified
batch. The distance is then predicted from the concantenated node embeddings of the two nodes.
# @package _global_
defaults:
- override /metrics:
- rmse
- override /decoder:
- edge_distance
dataset:
num_classes: 1
callbacks:
early_stopping:
monitor: val/edge_distance/rmse
mode: "min"
model_checkpoint:
monitor: val/edge_distance/rmse
mode: "min"
task:
task: "edge_distance_prediction"
transform:
_target_: proteinworkshop.tasks.edge_distance_prediction.EdgeDistancePredictionTransform
num_samples: 256
losses:
edge_distance: mse_loss
label_smoothing: 0.0
output:
- edge_distance
supervise_on:
- edge_distance
Auxiliary Tasks#
Auxiliary tasks define training objectives additional to the main training task. For instance, these can define auxiliary denoising objectives over sequence, coordinate or angle space.
Auxiliary tasks are implemented by modifying the experiment config to inject the additional metrics, decoders and losses.
Note
Aux tasks are specified with the following syntax:
python proteinworkshop/train.py ... +aux_task=nn_sequence
Sequence Denoising (nn_sequence)#
# Example:
python proteinworkshop/train.py dataset=cath encoder=gvp task=plddt_prediction +aux_task=nn_sequence
# @package _global_
defaults:
- _self_
- /decoder:
- residue_type
- /metrics@residue_type:
- /task@losses@residue_type: null
- /transforms:
- remove_missing_ca
- sequence_denoising
task:
losses:
residue_type: cross_entropy
output: ${oc.dict.keys:task.losses}
supervise_on: ${oc.dict.keys:task.losses}
aux_loss_coefficient:
residue_type: 0.1
Structure Denoising (nn_structure_r3)#
# Example:
python proteinworkshop/train.py dataset=cath encoder=gvp task=plddt_prediction +aux_task=nn_structure_r3
# @package _global_
defaults:
- _self_
- /decoder:
- pos_equiv
- /metrics: null
- /task@losses@pos: null
- /transforms:
- remove_missing_ca
- structure_denoising
task:
losses:
pos: mse_loss
output: ${oc.dict.keys:task.losses}
supervise_on: ${oc.dict.keys:task.losses}
aux_loss_coefficient:
pos: 0.1
Torsional Denoising (nn_structure_torsion)#
Noise is applied to the backbone torsion angles and Cartesian coordinates are recomputed using pNeRF and the uncorrupted bond lengths and angles prior to feature computation. Similarly to the coordinate denoising task, the model is then tasked with predicting either the per-residue angular noise or the original dihedral angles
Warning
This will subset the data to only include the backbone atoms
(N, Ca, C). The backbone oxygen can be placed with:
graphein.protein.tensor.reconstruction.place_fourth_coord().
This will break, for example, sidechain torsion angle computation for the first few chi angles that are partially defined by backbone atoms.
# Example:
python proteinworkshop/train.py dataset=cath encoder=gvp task=plddt_prediction +aux_task=nn_structure_torsion
# @package _global_
defaults:
- _self_
- /decoder:
- dihedrals
- /metrics: null
- /task@losses@pos: null
- /transforms:
- remove_missing_ca
- torsional_denoising
task:
losses:
dihedrals: mse_loss
output: ${oc.dict.keys:task.losses}
supervise_on: ${oc.dict.keys:task.losses}
aux_loss_coefficient:
pos: 0.1
Inverse Folding (inverse_folding)#
This adds an additional inverse folding objective to the model. I.e. the model is trained to predict the sequence of the input structure.
Warning
This will remove the residue-type node feature and sidechain torsion angles to avoid leakage of information from the target structure.
# Example:
python proteinworkshop/train.py dataset=cath encoder=gvp task=plddt_prediction +aux_task=inverse_folding
# @package _global_
defaults:
- _self_
- /decoder:
- residue_type
- /metrics@pos: null
- /task@losses@pos: null
- /transforms:
- remove_missing_ca
- inverse_folding
task:
losses:
residue_type: cross_entropy
output: ${oc.dict.keys:task.losses}
supervise_on: ${oc.dict.keys:task.losses}
None (none)#
# Example:
python proteinworkshop/train.py dataset=cath encoder=gvp task=plddt_prediction