Features#
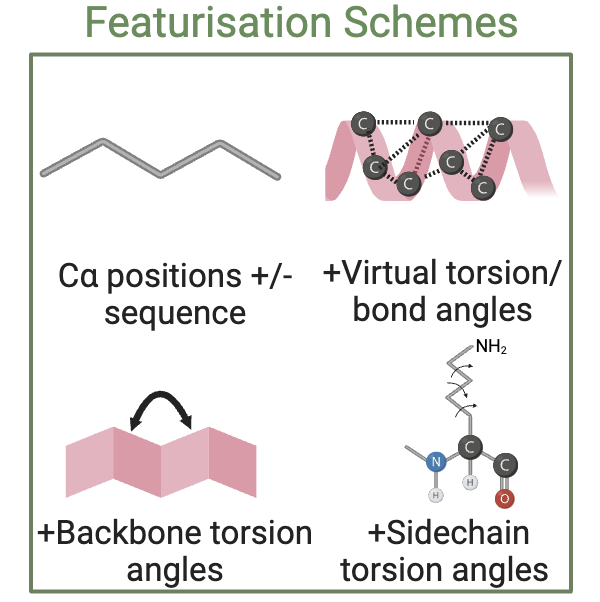
Part of the goal of the proteinworkshop benchmark is to investigate the impact of the degree to which increasing granularity of structural detail affects performance. To achieve this, we provide several featurisation schemes for protein structures.
Invariant Node Features#
N.B. All angular features are provided in [sin, cos] transformed form. E.g.: $textrm{dihedrals} = [sin(phi), cos(phi), sin(psi), cos(psi), sin(omega), cos(omega)]$, hence their dimensionality will be double the number of angles.
Name |
Description |
Dimensionality |
|---|---|---|
|
One-hot encoding of amino acid type |
21 |
|
Transformer-like positional encoding of sequence position |
16 |
|
Virtual torsion angle defined by four $Calpha$ atoms of residues $I{-1}, I, I{+1}, I{+2}$ |
2 |
|
Virtual bond angle (bend angle) defined by the three $Calpha$ atoms of residues $I{-2}, I, I_{+2}$ |
2 |
|
Backbone dihedral angles $(phi, psi, omega)$ |
6 |
|
Sidechain torsion angles $(chi_{1-4})$ |
8 |
Equivariant Node Features#
Name |
Description |
Dimensionality |
|---|---|---|
|
Forward and backward node orientation vectors (unit-normalized) |
2 |
Edge Construction#
We predominanty support two types of edges: $k$-NN and $epsilon$ edges.
Edge types can be specified as follows:
python proteinworkshop/train.py ... features.edge_types=[knn_16, knn_32, eps_16]
Where the suffix after knn or eps specifies $k$ (number of neighbours) or $epsilon$ (distance threshold in angstroms).
Invariant Edge Features#
Name |
Description |
Dimensionality |
|---|---|---|
|
Euclidean distance between source and target nodes |
1 |
|
Concatenated scalar node features of the source and target nodes |
Number of scalar node features $times 2$ |
|
Type annotation for each edge |
1 |
|
Sequence-based distance between source and target nodes |
1 |
|
Structured Transformer-inspired positional embedding of $i - j$ for source node $i$ and target node $j$ |
16 |
Equivariant Edge Features#
Name |
Description |
Dimensionality |
|---|---|---|
|
Edge directional vectors (unit-normalized) |
1 |
Default Features#
\(C_{\alpha}\) Only (ca_base)#
_target_: proteinworkshop.features.factory.ProteinFeaturiser
representation: CA
scalar_node_features:
- amino_acid_one_hot
vector_node_features: []
edge_types:
- knn_16
scalar_edge_features:
- edge_distance
vector_edge_features: []
\(C_{\alpha}\) + Sequence (ca_seq)#
_target_: proteinworkshop.features.factory.ProteinFeaturiser
representation: CA
scalar_node_features:
- amino_acid_one_hot
- sequence_positional_encoding
vector_node_features: []
edge_types:
- knn_16
scalar_edge_features:
- edge_distance
vector_edge_features: []
\(C_{\alpha}\) + Virtual Angles (ca_angles)#
_target_: proteinworkshop.features.factory.ProteinFeaturiser
representation: CA
scalar_node_features:
- amino_acid_one_hot
- sequence_positional_encoding
- alpha
- kappa
vector_node_features: []
edge_types:
- knn_16
scalar_edge_features:
- edge_distance
vector_edge_features: []
\(C_{\alpha}\) + Sequence + Backbone (ca_bb)#
_target_: proteinworkshop.features.factory.ProteinFeaturiser
representation: CA
scalar_node_features:
- amino_acid_one_hot
- sequence_positional_encoding
- alpha
- kappa
- dihedrals
vector_node_features: []
edge_types:
- knn_16
scalar_edge_features:
- edge_distance
vector_edge_features: []
\(C_{\alpha}\) + Sequence + Backbone + Sidechains (ca_sc)#
_target_: proteinworkshop.features.factory.ProteinFeaturiser
representation: CA
scalar_node_features:
- amino_acid_one_hot
- sequence_positional_encoding
- alpha
- kappa
- dihedrals
- sidechain_torsions
vector_node_features: []
edge_types:
- knn_16
scalar_edge_features:
- edge_distance
vector_edge_features: []